Myeloid leukemia vulnerabilities embedded in long noncoding RNA locus MYNRL15
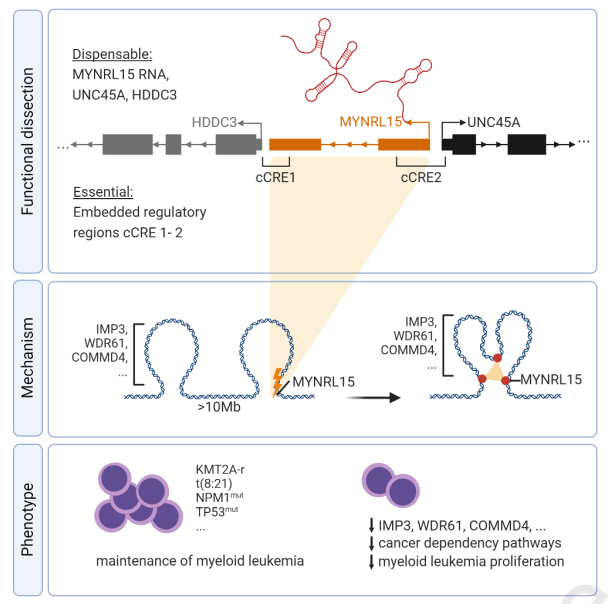
In this paper, we present the discovery of a functional noncoding locus on chromosome 15, which we named MYNRL15. The identification of this locus was led by exploring lncRNA loci in myeloid leukemia cell lines using CRISPRi-based dropout screens. Excision of MYNRL15 validated the proliferative disadvantage upon the loss of this locus. We also identified that the effects of the locus excision must occur in an RNA-independent manner. When focusing on the location of our lncRNA locus we found that MYNRL15 perturbation causes long range alterations on Chromosome 15 and that there are direct interaction sites found between YMNRL15 and sequences in a 500kb radius. Regarding protein coding effectors we identified specifically IMP3, WDR61, COMMD4 and SNUPN as being downregulated following MYNRL15 perturbation and contributing to the anti-leukemic effect of MYNRL15 targeting. Using primary AML patient cells in addition to patient derived xenografts we can highlight a selective impairment of AML cells underlining the therapeutic potential of MYNRL15 perturbation.