lncScape
A long noncoding RNA expression atlas of normal and malignant hematopoiesis
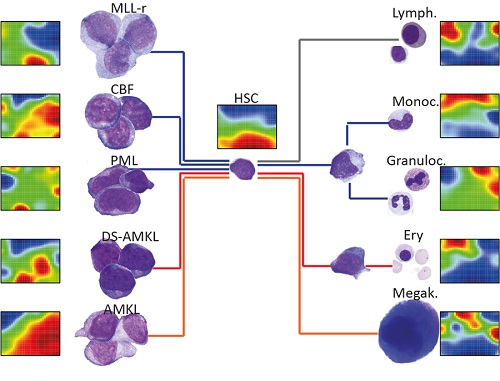
Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have emerged as crucial regulators of gene expression, epigenetics and cell fate decisions. To better define their roles in the blood system, we profiled miRNA, lncRNA and mRNA expression in human hematopoietic stem cells, granulocytes, monocytes, T-cells, B-cells, NK cells, megakaryocytes and erythroid cells. These ncRNA landscapes were compared with 46 pediatric AML patient samples, amounting to over 40,000 lncRNAs, 25,000 mRNAs and 900 miRNAs quantified on 146 arrays.
Clustering on lncRNAs and miRNAs structured our dataset into groups that match the input cell populations, underlining the cell type-specific expression patterns of ncRNA genes. Self-organizing maps also revealed gene sets that are coordinately upregulated in stem cells and pediatric AMLs. Via the lncScape online app, the expression of manifold miRNAs and lncRNAs can be retrieved across normal and malignant hematopoiesis. Our resource includes lineage-specific fingerprint lncRNAs, gene set enrichment analyses, and class prediction of co-expressed coding genes, so users can infer functions for every transcript.