CRISPRater
An integrated tool for predicting sgRNA efficacy in CRISPR-Cas9 experiments
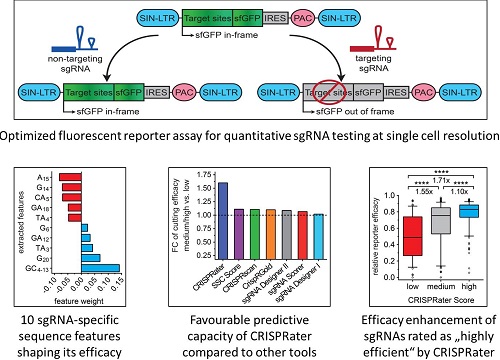
Genome editing with the clustered regularly interspaced short palindromic repeats (CRISPR) CRISPR-associated-9 (Cas9) technology has enabled unprecedented efficacy in reverse genetics and gene correction approaches. However, while both the reduction of off-target effects and the limitations of target site selection have been successfully tackled, the effort to eliminate variability in sgRNA efficacies – which affect experimental sensitivity – is still in its infancy.
With what is currently an unique approach involving the large-scale assessment of individual sgRNA efficacies at the single-cell level, and using widely-applied delivery tools in mammalian cells, we have developed the sgRNA prediction algorithm CRISPRater. Now integrated into the CCTop sgRNA scanning platform, CRISPRater offers an efficient way to prospectively increase genome editing performance and concurrently predict off-target sites (http://crispr.cos.uni-heidelberg.de/).