Browse through our online tools
Plasmids and more…
lncScape
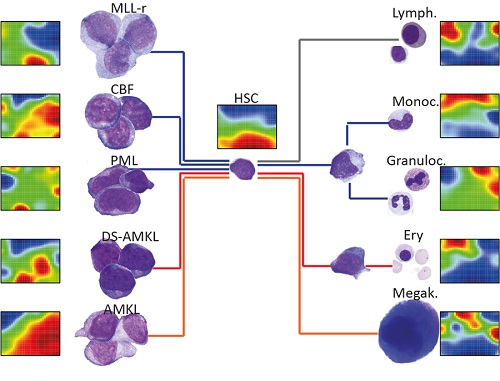
A long noncoding RNA expression atlas of normal and malignant hematopoiesis
Non-coding RNAs have emerged as crucial regulators of gene expression, epigenetics and cell fate decisions. To better define their role in hematopoiesis, we have profiled miRNA, lncRNA and mRNA expression in human hematopoietic stem cells and their differentiated progeny, including all the mature blood lineages. These ncRNA landscapes were compared with those of 46 pediatric AML patient samples, amounting to more than 40,000 lncRNAs, 25,000 mRNAs and 900 miRNAs quantified on 146 arrays.
Via the lncScape online app, the expression levels of manifold miRNAs and lncRNAs can be retrieved across normal and malignant hematopoiesis. Our resource includes lineage-specific fingerprint lncRNAs, gene set enrichment analyses, and class prediction of co-expressed coding genes, so that users can infer functions for every transcript.
Plasmids
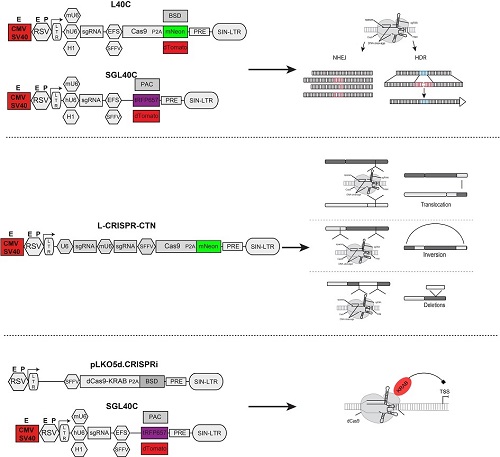
A catalog of advanced lentiviral CRISPR-Cas9 delivery vectors from our laboratory
The recent discovery and application of the CRISPR-Cas9 system has enabled unprecedented versatility and simplicity in genome editing. CRISPR-Cas9-based technologies are now used to inactivate genes, correct genetic mutations, induce large genomic deletions and chromosomal translocations, and even to control transcription and epigenetics. Our lab has developed a range of modular Cas9, sgRNA, and all-in one plasmids for efficient delivery of the CRISPR-Cas9 system into murine and human hematopoietic cells. We leverage these to model translocation-driven leukemiogenesis, to delineate the sequential acquisition of mutations during leukemia progression, to perform targeted gene correction, and to inhibit or activate the expression of non-coding RNAs. The sgRNA constructs can be used singly or multiplexed for large-scale forward-genetic screens.
CRISPRater
An integrated tool for predicting sgRNA efficacy in CRISPR-Cas9 experiments
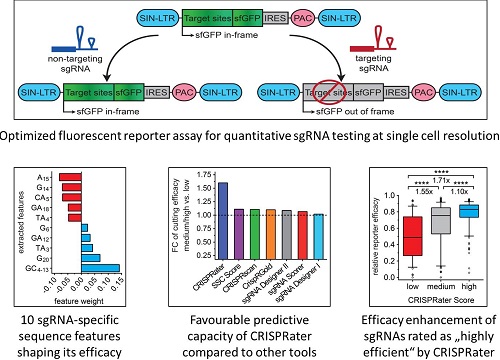
Genome editing with the clustered regularly interspaced short palindromic repeats (CRISPR) CRISPR-associated-9 (Cas9) technology has enabled unprecedented efficacy in reverse genetics and gene correction approaches. However, while both the reduction of off-target effects and the limitations of target site selection have been successfully tackled, the effort to eliminate variability in sgRNA efficacies – which affect experimental sensitivity – is still in its infancy.
With what is currently an unique approach involving the large-scale assessment of individual sgRNA efficacies at the single-cell level, and utilizing widely-applied delivery tools in mammalian cells, we have developed the sgRNA prediction algorithm CRISPRater (http://crispr.cos.uni-heidelberg.de/). Now integrated into the CCTop scanning platform, CRISPRater offers an efficient way to prospectively increase genome editing performance and concurrently predict off-target sites.