The non-coding RNA landscape of human hematopoiesis
The largest project in our lab for the last years was published today in Nature Communications. In the article, titled “The non-coding RNA landscape of human hematopoiesis and leukemia“, we illustrate non-coding RNA (ncRNA) expression profiles for human blood cells as well as for pediatric acute myeloid leukemia (AML) patient blasts, and provide a comprehensive resource for exploring the levels and potential functions of ncRNAs in normal and malignant hematopoiesis.
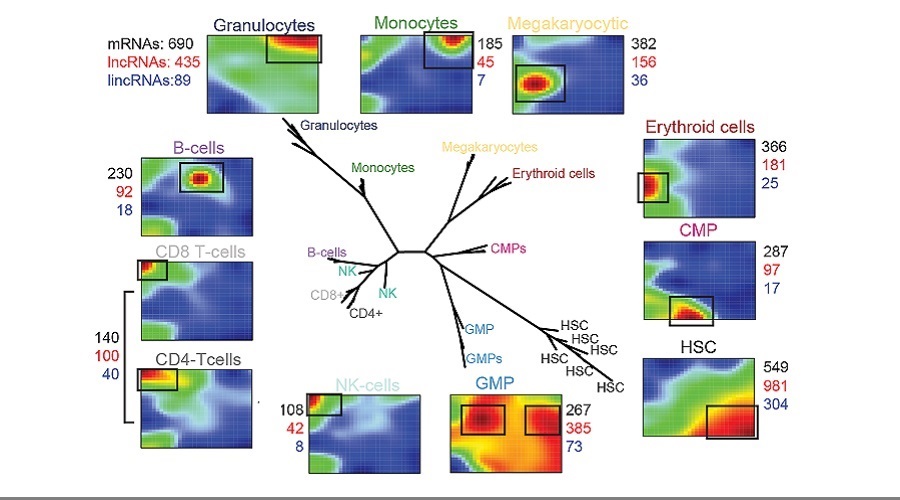
Despite the emergence of non-coding RNAs as crucial regulators of gene expression, their roles during normal and malignant hematopoesis remain incompletely understood. In particular, recent large-scale studies have highlighted the cell type- and developmental context-restricted expression patterns of long non-coding RNAs (lncRNAs), but functional studies have lagged behind these in silico discoveries. To address this knowledge gap, we established a ncRNA expression atlas of the human blood system covering ~40,000 lncRNAs, 20,000 mRNAs, and 900 microRNAs (miRNAs). Unique ncRNA expression profiles could be identified for every blood lineage, and a “guilt-by-association” approach involving co-expressed coding genes allowed the prediction of ncRNA functions.
One lncRNA identified by this pipeline, LINC00173, was gradually upregulated through the different stages granulopoiesis (i.e. from HSCs to CMPs to GMPs to granulocytes), and was predicted to act in concert with PRC2-associated networks. Indeed, we could show that LINC00173 controls myeloid differentiation, via direct association with PRC2 to silence “legacy” stem cell genes including the HOXA7-HOXA10 locus. We thus take this example as proof that our resource can guide hypothesis-driven studies on functional lncRNAs. Moreover, a multidimensional approach that integrated miRNA, lncRNA and positional data uncovered strong and coordinated upregulation of numerous ncRNAs within the DLK1-DIO3 locus on chromosome 14. Gain- and loss-of function studies in CD34+ HSPCs defined this locus as a novel positive regulator of megakaryopoiesis.
Last but not least, 46 pediatric AML patient samples were incorporated into the ncRNA expression atlas, to extend our resource and findings to malignant blood cells. Importantly, we discovered a core ncRNA stem cell signature in HSCs that was also upregulated in the AML samples, but absent from the mature lineages. In an independent cohort of 171 adult AML patients, high expression of the ncRNA stem cell signature predicted significantly better overall (OS) and event-free (EFS) survival. In contrast, downregulation of differentiation-associated ncRNAs predicted poor OS and EFS. Both the stem cell and differentiation-associated ncRNA signatures could further stratify patient prognoses based on a published 17-gene stemness score, demonstrating one possible aspect of the clinical value of ncRNAs.
Overall, these findings represent another step forward in the recent efforts to shed light on the non-coding transcriptome. In addition to providing a targeted, integrated expression resource across normal and malignant hematopoiesis, our article illustrates how these ncRNA landscapes can be used to inform greatly-needed downstream functional studies. Further, we provide evidence for the importance of ncRNAs in a clinical setting, highlighting their promise as potential prognostic indicators and/or therapeutic targets.